|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
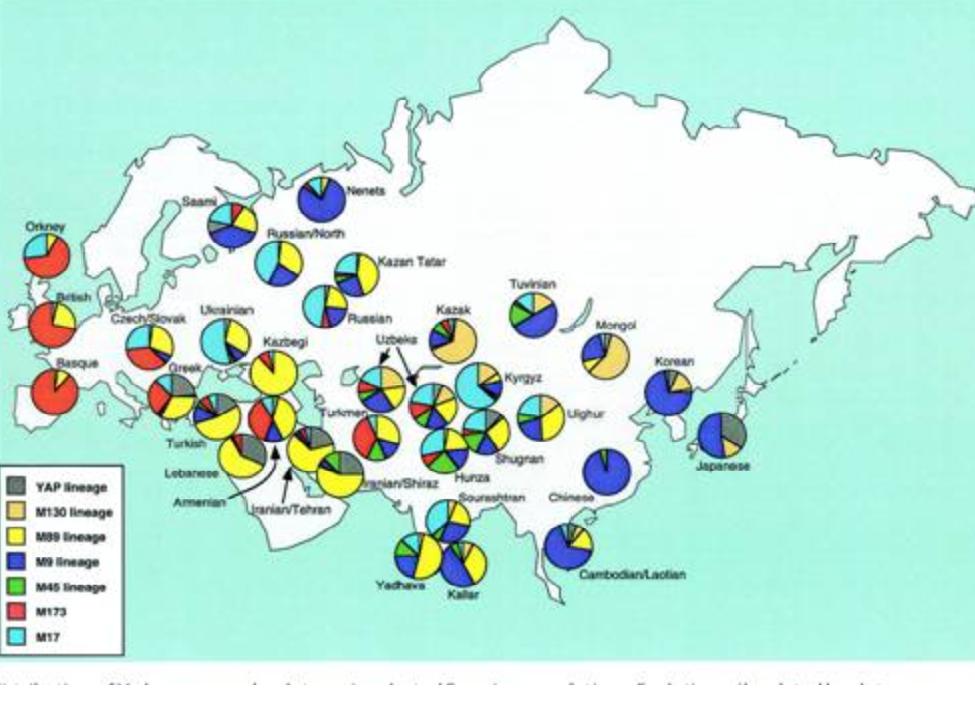
The haplogroup HG3 (or in the new nomenclature,
R1a1) is seen more frequently on the eastern side of Europe – for example, 9%
of the population of
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Haplogroup R1a The fabled haplogroup
R1a - or, more precisely, its subclade R1a1 - is
said to indicate a "Viking origin" when it
is found among men of British descent. This is the haplogroup
that will earn you a "Viking" certificate from Oxford Ancestors, and
its presence was the main focus of the Capelli
study "A Y Chromosome Census of The It is believed to have originated
among the Kurgan culture of western with
spreading the Indo-European languages to northwestern
pastoral economy, and to this day
their descendants bear the genetic traces of a dependence on livestock and animal
products. The incidence of milk tolerance among the Swedes, for instance, is
among the highest in the
world. The Scandinavians have long
believed that their ancestors originated in descent for the Jarls of Norway
from the warriors of Troy, and
the anthropologist Thor Heyerdahl - the author of
the classic "Kon Tiki" - spent his
final years attempting to trace the human originals of Wodin
and the Aesir back to R1a haplotypes
often score matches with Indians, Siberians, Chinese and other Asians - even
when they score no matches with
persons from of the
Silk Road, and their relationship with the people of haplogroup, and perhaps the less said about that,
the better. A person who does not belong to haplogroup R1a may, in fact, have a "deep
ancestry" in R1a accounts for only about 30%
of the men of about 35%,
and even R1b accounts for as much as 28%. Conversely, a person who does
belong to R1a does not
necessarily have Scandinavian ancestors - even if his people are from Suppose you take three Britons -
one whose grandfather was a Pakistani immigrant, another whose grandfather was a Polish pilot with the RAF,
and the third a Scot descended from one of the Hungarian noblemen who accompanied Margaret Atheling to
the court of Malcolm Canmore - and who, perhaps,
bears the name Drummond, Borthwick
or Crichton. All three gentlemen could easily
be R1a, but that doesn't make them Vikings. However, some
DNA genealogists have asserted not only that R1a was "Viking",
but that only R1a was truly Viking - and that
all the occurrence of R1b in the Norwegian population is due to the
importation of Celtic slaves. This is a curiously
Anglo-Centric argument. Vikings took slaves from many foreign lands, and sold
most of them to other
foreign lands. Most of the slaves in with
other Vikings. Even those foreign slaves who were imported to than
Celtic, as Slavs comprised the largest number of the slaves the Vikings
bought and sold. Slavs had been common
victims of the "peculiar institution" since Roman times. R1a is, in fact, far more
prevalent in counterargument that
it was not R1b, but R1a, whose incidence in R1a could also have entered Visigoths also settled in family of from a Visigothic family that obtained lands in Roman Gaul. Indo-Iranian nomads like the Alans and the Sarmatians also
probably carried R1a, and they found their way to Viking
DNA Among The Border Rivers Many of the fact that names and
Norse dialect, to archaeological finds of Viking artifacts
and "hogback" style tombstones. Most of these Vikings were actually Hiberno-Norse, which means that their forebears had
resided in and had
intermarried with the Irish Gaels. From the start of the wars in Boru
finally ejected the Vikings at the Battle of Clontarf
in 1014, there was a steady stream of refugees to the Isle of Man,
the Wirral (near Penrith,
which is in about 8 percent, according to the
Haplogroup R1a The fabled haplogroup R1a - or, more precisely, its subclade R1a1 - is said to indicate a "Viking
origin" when it
is found among men of British descent. This is the haplogroup
that will earn you a "Viking" certificate from Oxford Ancestors, and
its presence was the main focus of the Capelli
study "A Y Chromosome Census of The It is believed
to have originated among the Kurgan culture of western with
spreading the Indo-European languages to northwestern
pastoral
economy, and to this day their descendants bear the genetic traces of a
dependence on livestock and animal
products. The incidence of milk tolerance among the Swedes, for instance, is
among the highest in the
world. The
Scandinavians have long believed that their ancestors originated in descent for the
Jarls of Norway from the warriors of Troy, and
the anthropologist Thor Heyerdahl - the author of
the classic
"Kon Tiki" -
spent his final years attempting to trace the human originals of Wodin and the Aesir back to R1a haplotypes often score matches with Indians, Siberians,
Chinese and other Asians - even when they score no matches with
persons from of the
Silk Road, and their relationship with the people of haplogroup, and perhaps the less said about that,
the better. A person who
does not belong to haplogroup R1a may, in fact,
have a "deep ancestry" in R1a accounts for
only about 30% of the men of about 35%,
and even R1b accounts for as much as 28%. Conversely, a person who does
belong to R1a does not
necessarily have Scandinavian ancestors - even if his people are from Suppose you take
three Britons - one whose grandfather was a Pakistani immigrant, another
whose grandfather was a Polish
pilot with the RAF, and the third a Scot descended from one of the Hungarian
noblemen who accompanied
Margaret Atheling to the court of Malcolm Canmore -
and who, perhaps, bears the name Drummond, Borthwick or Crichton. All three
gentlemen could easily be R1a, but that doesn't make them Vikings. However, some
DNA genealogists have asserted not only that R1a was "Viking",
but that only R1a was truly Viking - and that
all the occurrence of R1b in the Norwegian population is due to the
importation of Celtic slaves. This is a curiously
Anglo-Centric argument. Vikings took slaves from many foreign lands, and sold
most of them to other
foreign lands. Most of the slaves in with
other Vikings. Even those foreign slaves who were imported to than
Celtic, as Slavs comprised the largest number of the slaves the Vikings
bought and sold. Slavs had been common
victims of the "peculiar institution" since Roman times. R1a is, in fact,
far more prevalent in counterargument that
it was not R1b, but R1a, whose incidence in R1a could also
have entered Visigoths also
settled in family of from a Visigothic family that obtained lands in Roman Gaul. Indo-Iranian
nomads like the Alans and the Sarmatians
also probably carried R1a, and they found their way to Viking
DNA Among The Border Reivers Many of the
Border Reiver families are rumored
to have Viking origin. That is a reasonable assumption in view of the fact that names and
Norse dialect, to archaeological finds of Viking artifacts
and "hogback" style tombstones. Most of these Vikings
were actually Hiberno-Norse, which means that their
forebears had resided in and had
intermarried with the Irish Gaels. From the start of the wars in Boru
finally ejected the Vikings at the Battle of Clontarf
in 1014, there was a steady stream of refugees to the Isle of Man,
the Wirral (near Penrith,
which is in about 8
percent, according to the Capelli study. The
percentage of R1a in our sample so far is less than half that. R1a Haplotype #16 Of the ten
highest frequencies for the haplotype below, all
but three fall in Bulgarian
gypsies, who may ultimately be of Indian origin, and in the Most of the
other areas where it is common in Fenno-Scandinavian
colonization, such as levels in Some have
speculated that Scandinavian R1a has a different geographical pattern from
Slavic R1a, in that the former has matches in Caucasus while
the latter is confined to pattern for
this haplotype bears a strong bias towards the
latter. Nonetheless, it
most likely came to
R1a Haplotype #21 This haplotype is widespread, but is clearly most common in
Eastern Europe, particularly The conventional
interpretation is to attribute any R1a haplotype
found in a person of British descent to the Norse Vikings, but
this geographical match pattern - at least in theory - could support an
ancestry among the Alans or the Sarmatians. However, far
more Danes and Norwegians are likely to have settled in Sarmatians, so
our final vote must go to the Scandinavians.
R1a
Haplotype #22 The match
pattern for this haplotype falls exclusively in It most likely
came to
R1a
Haplotype #25 The haplotype below exhibits a large number of hits in parts
of Saxony (e.g., and are
unusual for an R1a haplotype, and suggest an
Anglo-Danish origin. We would do well
to remember that the Angles, and the Jutes in particular, originated from
what is now known as also
served in It is
unrealistic to insist that all R1a in pre-Norman
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Haplogroup R1a
The fabled haplogroup R1a - or, more precisely, its subclade R1a1 - is said to indicate a "Viking
origin"
when it is
found among men of British descent. This is the haplogroup
that will earn you a "Viking" certificate
from Oxford Ancestors, and
its presence was the main focus of the Capelli study
"A Y Chromosome
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|

|
|
|
393 |
390 |
394 |
391 |
385a |
385b |
426 |
388 |
439 |
389i |
392 |
389ii |
458 |
459a |
459b |
455 |
454 |
447 |
437 |
448 |
449 |
464a |
464b |
464c |
464d |
460 |
gata |
yca |
yca |
456 |
607 |
576 |
570 |
cdy |
cdy |
442 |
438 |
|
|
|
|
|
19 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
h
4 |
ii a |
ii b |
|
|
|
|
a |
b |
|
|
|
Kit # |
loc1 |
loc2 |
loc3 |
loc4 |
loc5 |
loc6 |
loc7 |
loc8 |
loc9 |
loc10 |
loc11 |
loc12 |
loc13 |
loc14 |
loc15 |
loc16 |
loc17 |
loc18 |
loc19 |
loc20 |
loc21 |
loc22 |
loc23 |
loc24 |
Loc25 |
loc26 |
loc27 |
loc28 |
loc29 |
loc30 |
loc31 |
loc32 |
loc33 |
loc34 |
loc35 |
loc36 |
loc37 |
|
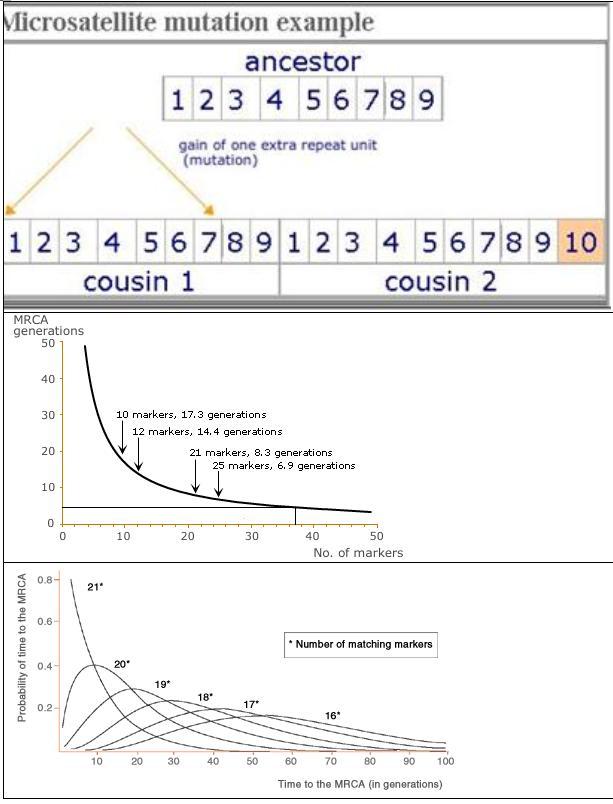
Using genetic markers, we can estimate
how closely people are related to each other. We do this by counting the
genetic differences (mutations) between two individuals.
The particular DNA markers we analyse
are called ‘short tandem repeats’, where small sections of the DNA code are
repeated several times. At any one of these short tandem repeats, the number of
repeats can increase or decrease, usually one at a time. Thus 9 repeats
of the code, GTCA, may suddenly be copied incorrectly within the body
and change to 10 repeats.
Many people who share a surname will
also share their haplotypes (i.e. have a 21/21
match). The graph below ('Matches against 21 markers') shows that
, mathematically, the most likely person to have your haplotype is zero generations away - i.e. you (look at the
line 21*). This if course makes perfect sense. But it
also means that as you increase the number of generations, the probability of
matching someone else becomes lower, which also makes sense. There is a higher
chance that mutations have occured.
If you match someone at 20 out of 21
markers, you'll get a slightly different probability curve. The most likely
MRCA is now not at zero generations, but further away.
Using 21 markers, it is usual for related individuals to share an exact haplotype i.e. a 21/21 match, although 20/21 and 19/21
matches should also be considered. Any more than this and the times to the MRCA
are just too long for a connection to be considered - as most surnames begun
much more recently
Haplogroup versus Haplotype versus Lineage
There is
a lot of talk about haplotype versus haplogroup. Some definitions from Peter de Knijff's paper "Messages through Bottlenecks: On the
Combined Use of Slow and Fast Evolving Polymorphic Markers on the Human Y Chromosome" [Am. J. Hum. Genetics, 67:1055-1061
(2000)]. "Distinct Y chromosomes identified by STR's
are designated as 'haplotypes.'" Stated in
another way: a haplotype is represented by the number
of repeats at certain alleles, or markers on the y
chromosome. "Distinct Y chromosomes, defined solely on the basis of
(unique mutation events)
Care
must be taken when comparing results. There is always a possibility that
someone may be comparing a haplotype of someone in a
different haplogroup. In one-name studies this
"threat" is diminished. However, if you are searching databases that
have haplotypes, you have to be sure that you are
comparing apples and apples, not apples and oranges--comparing those in your
own haplogroup (which these databases do not yet
provide for). While I thought that future testing should include more biallelic polymorphisms in order to determine haplogroup, Dr. Mark Jobling says
what is needed are more microsatellites in testing,
not biallelic polymorphisms. At present we are seeing
people compare apples and oranges, saying that they may have a common ancestor
with some other surname such as Duerinck, when in
reality, we both may be in different haplogroups. I
am currently reading 3 papers from the American Journal of Human Genetics on
these issues: "Y-Chromosomal Diversity in Europe is Clinal
and Influenced Primarily by Geography, Rather than by Language" by Rosser,
et al. [AJHG vol 67 page 1526 (2000)];
"Estimating Scandinavian and Gaelic Ancestry in the Male Settlers" by
Helgason et al. [AJHG vol
67 page 697 (2000)]; and "Messages through Bottlenecks: On the Combined
Use of Slow and Fast Evolving Polymorphic Markers on the Human Y
Chromosome" by de Knijff [AJHG vol 67 page 1055 (2000)].
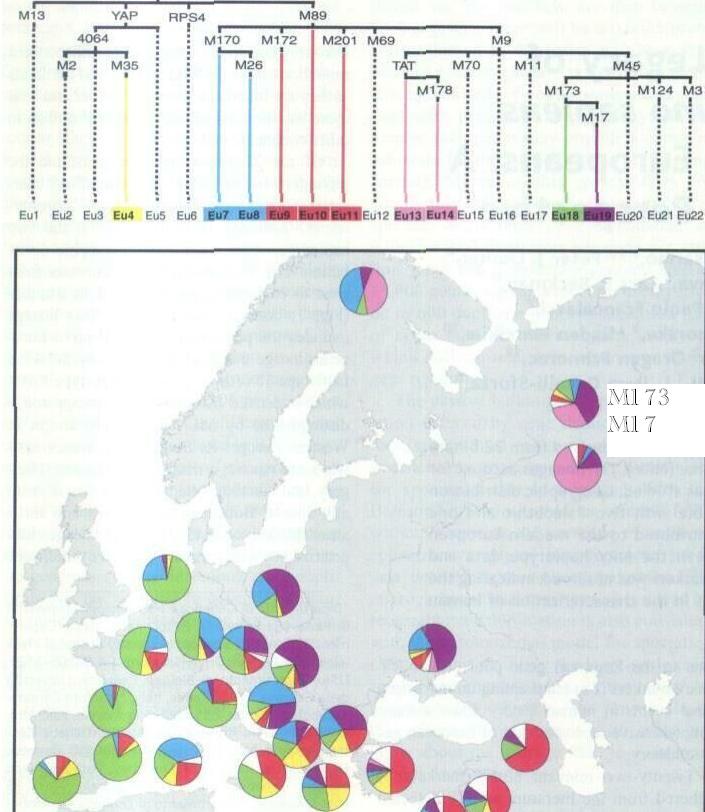
It appears that HG 1 may be
the main pre-Ice Age male element in western Europe
(for y-chromosome analysis), similar to Prof. Sykes's Daughters of Eve (mtDNA analysis). HUGO, the gene nomenclature body, and the
University College of London would like to resolve the differences in
nomenclature for the haplogroups.
Most Recent Common
Ancestor (MRCA)
In order to understand
MRCA, you must understand the following:
--MRCA (most recent common ancestor): the common ancestor between 2 people.
--generation: it means every 20 years here (lot of discussion on this
point--most of us are inclined to believe that 25 years is a better estimate).
--Heyer principle: y
chromosome mutations occur generally once every
500 generations per locus (or "per marker") (mutation rate of
0.2% or 0.002, Heyer et al. 1997)
--Example of Heyer principle: Using the Heyer principle, we would expect EACH locus to change 1
digit in 500 generations. For example, we would expect that a 5 (repeat units)
in locus #1 would move to either a 4 or a
So, instead of ONE locus, how about looking at 12 loci (or 15
or 20 etc.)? If we use a 12 marker test,
in 500 (generations) / 12 (markers) = 41.667 generations we would expect
to have a change in 1 of the 12 markers by 1 digit. OR, to be in the
"club" of Duerincks, if the Common Ancestor
was farther back in time than 41.667 generations (a generation = 20 years),
then it would be totally expected that a "real" related Duerinck (Kit
Getting back to the reasonableness
of expecting a mutation to occur every 41 or so generations if a 12 marker test
(testing 12 loci) is used, what does the mutation rate become if more markers
are used? Simple. You must also remember that loci
changes (mutations) can occur at any time. Using the y chromosome mutation rate
PER MARKER of once every 500 generations, if 15 loci are measured, 500 divided
by 15 = every 30 generations there could be a mutation at a loci. If 17 loci
are measured, 500 divided by 17 = every 29 generations. At 20 loci being
measured, 500 divided by 20 = every 25 generations there could be a mutation at
a loci. Where 21 loci are measured and you have a 21/21 match, this indicates
that there is a 50% probability that the MRCA is within 9 generations and a 90%
probability that the MRCA is within 28 generations.
As genealogists we want to
know when the MRCA between two people lived. We know that where all 12 markers
match, there is a 50% probability that the MRCA was no longer than 14.5
generations (290 years), and a 90% probability that the MRCA was within the
last 48 generations (63 generations at 95%). The range for a 12/12 match is 1
to 63 generations. Further, if a test uses 12 different loci (a 12 marker
test), it is reasonable to expect a change to occur every 41 or so generations;
however, these changes can take place at any time.
What if there is an 11/12
marker match? For an 11/12 match, there is a 50% probability that the
MRCA lived no longer than 36 generations. This translates into 36 x 20 = 720
years. The range for an 11/12 match is 1 to 104 generations (85 generations at
90%, 104 at 95%).
What if there is a 10/12
match? For a 10/12 match, there is a 50% probability that the MRCA
lived no longer than 61 generations. The range for a 10/12 match is 1 to 145 generations
(122 at 90%, 145 at 95% probability).
We do know that if the
results of 2 people are different by 1 mutation, that these people are related.
If different by 2 mutations, probably related. Is
there a difference if there is a 2 repeat value change on one loci OR if there is are 1 repeat value changes on two loci?
As we see 3 or more mutations, the people who may be related are very distantly
related. The concept of MRCA puts the relation back many generations, more like
1,000 to 2,000 years for 3 mutations. For 3 or
more mutations out of a 12 marker test, people are not related from a
genealogical standpoint.
Just an update when using
21 markers: if you match 21/21 then the likelihood of having a common ancestor
with that person is that in 50% of the cases the common ancestor would have
been within 8.3 generations. Of course it could be more recent and it could be
farther back.
At a 25 marker match
between 2 individual males (with the same or variant surname), there is a 50%
probability that the most recent common ancestor is 7 generations back or less.
We are speaking of confidence intervals. In other words, 50% will find their
common ancestor within that range. If you want to be more confident (think of
it as placing a gambling bet--when did the MRCA live?), the number of
generations back to the MRCA would increase. How confident are you at 95%
probability? 95% will find their common ancestor within 30 generations ago or
less for a 25/25 marker match. You can find the MRCA within the range of 0 to 37
generations for a 25/25 marker match.
Did you ever wonder what
DYS stands for? D = DNA, Y = Y-chromosome, S = (unique) segment [or
"single copy sequence" according to "Forensic DNA Typing"
by John Butler]. The DYS numbering scheme (e.g. DYS388, DYS390) for the Y-STR haplotype loci is controlled and administered by an
international standards body called HUGO Human Gene Nomenclature Committee
based at
PHYLOGENETIC TREE
Here are most of the
results represented in a phylogenetic tree. This
graph was done by Ann Turner, GENEALOGY-DNA List Administrator, and is used
here with her express permission. You will note the markers are DYS markers for
the known markers and FT markers for those markers used by FTDNA that have not
yet been made public. The software that was used is available at Fluxus-Engineering. The instruction page notes that in
human Y STRs, compound STRs
such as DYS389II should be resolved into its mutational subcomponents m, n, and
q to avoid artefacts. In the graph below, the Reduced Median (RM) network
method was used.

|
|
|
|
The above phylogenetic tree shows 2 clusters (tree needs to be
updated showing closer relationships by eliminating an error in locus 19 which
allele was previously 12 but has now been corrected to
The second cluster, at the
top of the graph, shows participants 299 Von Duering,
297 Dieringer, and 337 Duering
to be more closely related than those participants in the first cluster and the
bottom. Participants 283 and 287 were not graphed because they were the
furthest away from any of the other participants and also because the software
used started messing up.
http://www.cstl.nist.gov/div831/strbase/ppt/ISFG2001.ppt
- 16
http://wanclik.free.fr/family_tree.htm